class: clear background-image: url(fig/Slide1.PNG) background-size: 990px <style type="text/css"> h2 { color: brown; } </style> --- class: clear background-image: url(fig/Slide2.PNG) background-size: 850px --- class: clear background-image: url(fig/Slide3.PNG) background-size: 850px --- class: clear background-image: url(fig/Slide4.PNG) background-size: 850px --- class: clear background-image: url(fig/Slide5.PNG) background-size: 850px --- class: clear background-image: url(fig/Slide6.PNG) background-size: 850px --- class: clear background-image: url(fig/Slide7.PNG) background-size: 850px --- class: clear background-image: url(fig/Slide8.PNG) background-size: 850px --- class: clear background-image: url(fig/Slide9.PNG) background-size: 850px --- class: clear background-image: url(fig/Slide10.PNG) background-size: 850px --- class: clear background-image: url(fig/Slide11.PNG) background-size: 850px --- class: clear background-image: url(fig/Slide12.PNG) background-size: 850px --- # PCA no R ## Instalar os pacotes necessários a execução da PCA ```r install.packages("vegan") install.packages("ggfortify") install.packages("psych") install.packages("MVN") install.packages("tidyverse") intall.packages("kableExtra") intall.packages("GGally") ``` ## Carregando os pacotes ```r library(vegan) library(ggfortify) library(psych) library(MVN) library(tidyverse) library(kableExtra) library(GGally) ``` --- # PCA no R ## Abrir o banco de dados ```r ## Abrir a base de dados dados = read.csv2("PCA.csv", sep=",", dec=".", header=TRUE, strip.white=TRUE) ## Dados sem a linha nominal dados1 = dados[,-1] attach(dados) dados1 ``` <div style="width: 800px; height: 350px; white-space: nowrap; overflow-x: scroll; overflow-y: scroll; border: 0; padding: 0px; display: inline-block;"> <table class=" lightable-classic" style="font-family: Cambria; margin-left: auto; margin-right: auto;"> <thead> <tr> <th style="text-align:right;"> Temperatura </th> <th style="text-align:right;"> pH </th> <th style="text-align:right;"> Potêncial.de.oxirredução </th> <th style="text-align:right;"> Turbidez </th> <th style="text-align:right;"> Oxigênio.dissolvido </th> <th style="text-align:right;"> Espécie.A </th> <th style="text-align:right;"> Espécie.B </th> </tr> </thead> <tbody> <tr> <td style="text-align:right;"> 18.0 </td> <td style="text-align:right;"> 6.30 </td> <td style="text-align:right;"> 213 </td> <td style="text-align:right;"> 10.60 </td> <td style="text-align:right;"> 10.03 </td> <td style="text-align:right;"> 10 </td> <td style="text-align:right;"> 10 </td> </tr> <tr> <td style="text-align:right;"> 17.0 </td> <td style="text-align:right;"> 7.11 </td> <td style="text-align:right;"> 143 </td> <td style="text-align:right;"> 7.11 </td> <td style="text-align:right;"> 9.00 </td> <td style="text-align:right;"> 9 </td> <td style="text-align:right;"> 9 </td> </tr> <tr> <td style="text-align:right;"> 17.5 </td> <td style="text-align:right;"> 6.70 </td> <td style="text-align:right;"> 311 </td> <td style="text-align:right;"> 13.70 </td> <td style="text-align:right;"> 11.84 </td> <td style="text-align:right;"> 8 </td> <td style="text-align:right;"> 8 </td> </tr> <tr> <td style="text-align:right;"> 18.6 </td> <td style="text-align:right;"> 6.00 </td> <td style="text-align:right;"> 347 </td> <td style="text-align:right;"> 22.40 </td> <td style="text-align:right;"> 11.00 </td> <td style="text-align:right;"> 7 </td> <td style="text-align:right;"> 7 </td> </tr> <tr> <td style="text-align:right;"> 19.0 </td> <td style="text-align:right;"> 7.00 </td> <td style="text-align:right;"> 246 </td> <td style="text-align:right;"> 20.30 </td> <td style="text-align:right;"> 9.75 </td> <td style="text-align:right;"> 6 </td> <td style="text-align:right;"> 6 </td> </tr> <tr> <td style="text-align:right;"> 17.2 </td> <td style="text-align:right;"> 7.20 </td> <td style="text-align:right;"> 365 </td> <td style="text-align:right;"> 8.46 </td> <td style="text-align:right;"> 9.77 </td> <td style="text-align:right;"> 5 </td> <td style="text-align:right;"> 5 </td> </tr> <tr> <td style="text-align:right;"> 17.0 </td> <td style="text-align:right;"> 6.80 </td> <td style="text-align:right;"> 89 </td> <td style="text-align:right;"> 17.60 </td> <td style="text-align:right;"> 7.00 </td> <td style="text-align:right;"> 4 </td> <td style="text-align:right;"> 4 </td> </tr> <tr> <td style="text-align:right;"> 18.0 </td> <td style="text-align:right;"> 7.12 </td> <td style="text-align:right;"> 64 </td> <td style="text-align:right;"> 5.56 </td> <td style="text-align:right;"> 10.90 </td> <td style="text-align:right;"> 11 </td> <td style="text-align:right;"> 11 </td> </tr> <tr> <td style="text-align:right;"> 18.9 </td> <td style="text-align:right;"> 7.00 </td> <td style="text-align:right;"> 123 </td> <td style="text-align:right;"> 16.20 </td> <td style="text-align:right;"> 9.70 </td> <td style="text-align:right;"> 12 </td> <td style="text-align:right;"> 12 </td> </tr> <tr> <td style="text-align:right;"> 17.4 </td> <td style="text-align:right;"> 6.90 </td> <td style="text-align:right;"> 264 </td> <td style="text-align:right;"> 11.50 </td> <td style="text-align:right;"> 11.38 </td> <td style="text-align:right;"> 13 </td> <td style="text-align:right;"> 13 </td> </tr> <tr> <td style="text-align:right;"> 18.1 </td> <td style="text-align:right;"> 6.80 </td> <td style="text-align:right;"> 167 </td> <td style="text-align:right;"> 10.80 </td> <td style="text-align:right;"> 12.74 </td> <td style="text-align:right;"> 14 </td> <td style="text-align:right;"> 14 </td> </tr> <tr> <td style="text-align:right;"> 17.0 </td> <td style="text-align:right;"> 6.70 </td> <td style="text-align:right;"> 156 </td> <td style="text-align:right;"> 6.00 </td> <td style="text-align:right;"> 7.84 </td> <td style="text-align:right;"> 12 </td> <td style="text-align:right;"> 12 </td> </tr> <tr> <td style="text-align:right;"> 20.0 </td> <td style="text-align:right;"> 7.80 </td> <td style="text-align:right;"> 116 </td> <td style="text-align:right;"> 12.70 </td> <td style="text-align:right;"> 10.73 </td> <td style="text-align:right;"> 2 </td> <td style="text-align:right;"> 6 </td> </tr> <tr> <td style="text-align:right;"> 21.0 </td> <td style="text-align:right;"> 7.00 </td> <td style="text-align:right;"> 141 </td> <td style="text-align:right;"> 12.50 </td> <td style="text-align:right;"> 9.29 </td> <td style="text-align:right;"> 5 </td> <td style="text-align:right;"> 5 </td> </tr> <tr> <td style="text-align:right;"> 19.6 </td> <td style="text-align:right;"> 9.00 </td> <td style="text-align:right;"> 238 </td> <td style="text-align:right;"> 19.40 </td> <td style="text-align:right;"> 10.02 </td> <td style="text-align:right;"> 4 </td> <td style="text-align:right;"> 4 </td> </tr> <tr> <td style="text-align:right;"> 18.8 </td> <td style="text-align:right;"> 8.00 </td> <td style="text-align:right;"> 245 </td> <td style="text-align:right;"> 9.06 </td> <td style="text-align:right;"> 8.76 </td> <td style="text-align:right;"> 6 </td> <td style="text-align:right;"> 11 </td> </tr> <tr> <td style="text-align:right;"> 21.0 </td> <td style="text-align:right;"> 8.10 </td> <td style="text-align:right;"> 301 </td> <td style="text-align:right;"> 6.00 </td> <td style="text-align:right;"> 9.49 </td> <td style="text-align:right;"> 6 </td> <td style="text-align:right;"> 12 </td> </tr> <tr> <td style="text-align:right;"> 21.6 </td> <td style="text-align:right;"> 8.00 </td> <td style="text-align:right;"> 299 </td> <td style="text-align:right;"> 13.50 </td> <td style="text-align:right;"> 10.51 </td> <td style="text-align:right;"> 7 </td> <td style="text-align:right;"> 13 </td> </tr> <tr> <td style="text-align:right;"> 22.0 </td> <td style="text-align:right;"> 8.00 </td> <td style="text-align:right;"> 100 </td> <td style="text-align:right;"> 6.44 </td> <td style="text-align:right;"> 8.87 </td> <td style="text-align:right;"> 1 </td> <td style="text-align:right;"> 14 </td> </tr> <tr> <td style="text-align:right;"> 21.8 </td> <td style="text-align:right;"> 8.10 </td> <td style="text-align:right;"> 95 </td> <td style="text-align:right;"> 10.40 </td> <td style="text-align:right;"> 12.71 </td> <td style="text-align:right;"> 2 </td> <td style="text-align:right;"> 10 </td> </tr> <tr> <td style="text-align:right;"> 20.0 </td> <td style="text-align:right;"> 7.90 </td> <td style="text-align:right;"> 125 </td> <td style="text-align:right;"> 8.96 </td> <td style="text-align:right;"> 8.56 </td> <td style="text-align:right;"> 5 </td> <td style="text-align:right;"> 9 </td> </tr> <tr> <td style="text-align:right;"> 20.7 </td> <td style="text-align:right;"> 8.90 </td> <td style="text-align:right;"> 130 </td> <td style="text-align:right;"> 10.90 </td> <td style="text-align:right;"> 9.76 </td> <td style="text-align:right;"> 6 </td> <td style="text-align:right;"> 8 </td> </tr> <tr> <td style="text-align:right;"> 20.7 </td> <td style="text-align:right;"> 8.50 </td> <td style="text-align:right;"> 92 </td> <td style="text-align:right;"> 13.40 </td> <td style="text-align:right;"> 8.05 </td> <td style="text-align:right;"> 7 </td> <td style="text-align:right;"> 7 </td> </tr> <tr> <td style="text-align:right;"> 22.8 </td> <td style="text-align:right;"> 8.40 </td> <td style="text-align:right;"> 133 </td> <td style="text-align:right;"> 9.01 </td> <td style="text-align:right;"> 7.81 </td> <td style="text-align:right;"> 9 </td> <td style="text-align:right;"> 5 </td> </tr> <tr> <td style="text-align:right;"> 25.0 </td> <td style="text-align:right;"> 8.60 </td> <td style="text-align:right;"> 150 </td> <td style="text-align:right;"> 17.00 </td> <td style="text-align:right;"> 10.27 </td> <td style="text-align:right;"> 10 </td> <td style="text-align:right;"> 9 </td> </tr> <tr> <td style="text-align:right;"> 24.0 </td> <td style="text-align:right;"> 8.50 </td> <td style="text-align:right;"> 239 </td> <td style="text-align:right;"> 26.60 </td> <td style="text-align:right;"> 9.63 </td> <td style="text-align:right;"> 9 </td> <td style="text-align:right;"> 3 </td> </tr> <tr> <td style="text-align:right;"> 23.0 </td> <td style="text-align:right;"> 8.70 </td> <td style="text-align:right;"> 197 </td> <td style="text-align:right;"> 23.40 </td> <td style="text-align:right;"> 13.30 </td> <td style="text-align:right;"> 8 </td> <td style="text-align:right;"> 2 </td> </tr> <tr> <td style="text-align:right;"> 24.5 </td> <td style="text-align:right;"> 8.40 </td> <td style="text-align:right;"> 188 </td> <td style="text-align:right;"> 26.70 </td> <td style="text-align:right;"> 7.76 </td> <td style="text-align:right;"> 7 </td> <td style="text-align:right;"> 1 </td> </tr> <tr> <td style="text-align:right;"> 23.8 </td> <td style="text-align:right;"> 8.40 </td> <td style="text-align:right;"> 288 </td> <td style="text-align:right;"> 15.00 </td> <td style="text-align:right;"> 9.04 </td> <td style="text-align:right;"> 6 </td> <td style="text-align:right;"> 2 </td> </tr> <tr> <td style="text-align:right;"> 25.0 </td> <td style="text-align:right;"> 8.30 </td> <td style="text-align:right;"> 255 </td> <td style="text-align:right;"> 21.30 </td> <td style="text-align:right;"> 9.71 </td> <td style="text-align:right;"> 5 </td> <td style="text-align:right;"> 1 </td> </tr> <tr> <td style="text-align:right;"> 25.7 </td> <td style="text-align:right;"> 7.90 </td> <td style="text-align:right;"> 204 </td> <td style="text-align:right;"> 14.40 </td> <td style="text-align:right;"> 9.66 </td> <td style="text-align:right;"> 4 </td> <td style="text-align:right;"> 2 </td> </tr> <tr> <td style="text-align:right;"> 25.9 </td> <td style="text-align:right;"> 9.00 </td> <td style="text-align:right;"> 224 </td> <td style="text-align:right;"> 17.60 </td> <td style="text-align:right;"> 8.36 </td> <td style="text-align:right;"> 11 </td> <td style="text-align:right;"> 3 </td> </tr> <tr> <td style="text-align:right;"> 24.0 </td> <td style="text-align:right;"> 9.10 </td> <td style="text-align:right;"> 188 </td> <td style="text-align:right;"> 17.80 </td> <td style="text-align:right;"> 9.99 </td> <td style="text-align:right;"> 12 </td> <td style="text-align:right;"> 0 </td> </tr> <tr> <td style="text-align:right;"> 23.0 </td> <td style="text-align:right;"> 9.20 </td> <td style="text-align:right;"> 275 </td> <td style="text-align:right;"> 26.90 </td> <td style="text-align:right;"> 8.01 </td> <td style="text-align:right;"> 13 </td> <td style="text-align:right;"> 1 </td> </tr> <tr> <td style="text-align:right;"> 28.9 </td> <td style="text-align:right;"> 9.00 </td> <td style="text-align:right;"> 216 </td> <td style="text-align:right;"> 22.00 </td> <td style="text-align:right;"> 7.26 </td> <td style="text-align:right;"> 14 </td> <td style="text-align:right;"> 2 </td> </tr> <tr> <td style="text-align:right;"> 25.0 </td> <td style="text-align:right;"> 8.90 </td> <td style="text-align:right;"> 197 </td> <td style="text-align:right;"> 14.10 </td> <td style="text-align:right;"> 10.04 </td> <td style="text-align:right;"> 12 </td> <td style="text-align:right;"> 0 </td> </tr> </tbody> </table> --- # PCA no R ## Pressupostos para a aplicação da Análise de componentes principais ### Todas as variáveis são númericas? ```r str(dados1) ``` ``` ## 'data.frame': 36 obs. of 7 variables: ## $ Temperatura : num 18 17 17.5 18.6 19 17.2 17 18 18.9 17.4 ... ## $ pH : num 6.3 7.11 6.7 6 7 7.2 6.8 7.12 7 6.9 ... ## $ Potêncial.de.oxirredução: int 213 143 311 347 246 365 89 64 123 264 ... ## $ Turbidez : num 10.6 7.11 13.7 22.4 20.3 8.46 17.6 5.56 16.2 11.5 ... ## $ Oxigênio.dissolvido : num 10.03 9 11.84 11 9.75 ... ## $ Espécie.A : int 10 9 8 7 6 5 4 11 12 13 ... ## $ Espécie.B : int 10 9 8 7 6 5 4 11 12 13 ... ``` --- # PCA no R ## Pressupostos para a aplicação da Análise de componentes principais ### Associação entre variáveis <div style="width: 800px; height: 480px; white-space: nowrap; overflow-x: scroll; overflow-y: scroll; border: 0; padding: 0px; display: inline-block;"> ```r cor = cor(dados1) ggpairs(cbind(dados1)) ``` 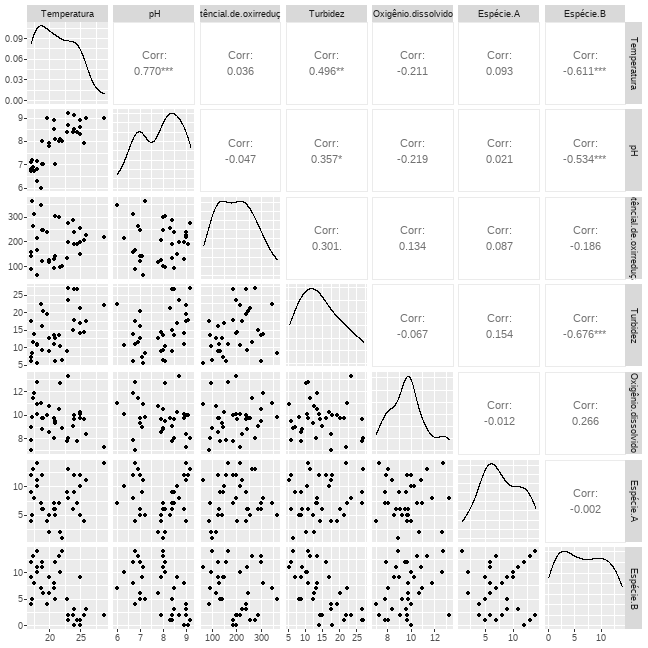<!-- --> --- # PCA no R ## Pressupostos para a aplicação da Análise de componentes principais ### Associação entre variáveis **Teste de esferécidade de Bartlett**: Teste de esfericidade de bartlett entre a matriz de correlação (cor) e o tamanho da amostra (n). Ele apresenta a significância da associação entre pelo menos algumas das variáveis amostradas (P<0,05). ```r bartlett.test(dados1) ``` ``` ## ## Bartlett test of homogeneity of variances ## ## data: dados1 ## Bartlett's K-squared = 925.76, df = 6, p-value < 2.2e-16 ``` --- # PCA no R ## Pressupostos para a aplicação da Análise de componentes principais ### Cosistência dos dados **Critério de Kayser-Meyer-Olkin (KMO)**: Indica a proporção da variância dos dados que pode ser considerada comum a todas as variáveis. Quanto mais próximo de 1 melhor o resultado. - \> 0,9 Excelente - (0,8; 0,9] Meritória - (0,7; 0,8] Intermedia - (0,6; 0,7] Medíocre - (0,5; 0,6] Mísera - < 0,5 Inaceitável <div style="width: 800px; height: 300px; white-space: nowrap; overflow-x: scroll; overflow-y: scroll; border: 0; padding: 0px; display: inline-block;"> ```r KMO(cor) ``` ``` ## Kaiser-Meyer-Olkin factor adequacy ## Call: KMO(r = cor) ## Overall MSA = 0.69 ## MSA for each item = ## Temperatura pH Potêncial.de.oxirredução ## 0.71 0.68 0.63 ## Turbidez Oxigênio.dissolvido Espécie.A ## 0.69 0.64 0.35 ## Espécie.B ## 0.73 ``` --- # PCA no R ## Pressupostos para a aplicação da Análise de componentes principais ### Teste de normalidade multivariada (mais informações sobre a função `mvn` [aqui](http://127.0.0.1:8746/help/library/MVN/doc/MVN.pdf)) <div style="width: 800px; height: 450px; white-space: nowrap; overflow-x: scroll; overflow-y: scroll; border: 0; padding: 0px; display: inline-block;"> ```r mvn(dados1) ``` ``` ## $multivariateNormality ## Test Statistic p value Result ## 1 Mardia Skewness 73.2150300351476 0.793488331389028 YES ## 2 Mardia Kurtosis -1.45240061170039 0.146390246329282 YES ## 3 MVN <NA> <NA> YES ## ## $univariateNormality ## Test Variable Statistic p value Normality ## 1 Shapiro-Wilk Temperatura 0.9446 0.0707 YES ## 2 Shapiro-Wilk pH 0.9370 0.0409 NO ## 3 Shapiro-Wilk Potêncial.de.oxirredução 0.9726 0.4993 YES ## 4 Shapiro-Wilk Turbidez 0.9500 0.1045 YES ## 5 Shapiro-Wilk Oxigênio.dissolvido 0.9648 0.3016 YES ## 6 Shapiro-Wilk Espécie.A 0.9620 0.2469 YES ## 7 Shapiro-Wilk Espécie.B 0.9404 0.0524 YES ## ## $Descriptives ## n Mean Std.Dev Median Min Max 25th ## Temperatura 36 21.152778 3.1360108 20.850 17.00 28.9 18.475 ## pH 36 7.870278 0.8981043 8.000 6.00 9.2 7.000 ## Potêncial.de.oxirredução 36 197.888889 78.1346010 197.000 64.00 365.0 132.250 ## Turbidez 36 14.591667 6.1847825 13.600 5.56 26.9 10.065 ## Oxigênio.dissolvido 36 9.681667 1.5087848 9.705 7.00 13.3 8.710 ## Espécie.A 36 7.833333 3.5576879 7.000 1.00 14.0 5.000 ## Espécie.B 36 6.694444 4.3936281 6.500 0.00 14.0 2.750 ## 75th Skew Kurtosis ## Temperatura 23.850 0.38700260 -0.8576499 ## pH 8.525 -0.28339071 -1.2046862 ## Potêncial.de.oxirredução 248.250 0.24397736 -0.9379144 ## Turbidez 18.200 0.42657020 -0.8655783 ## Oxigênio.dissolvido 10.330 0.45935368 -0.1873903 ## Espécie.A 11.000 0.07011761 -1.0521418 ## Espécie.B 10.250 0.09149470 -1.3697805 ``` --- # PCA no R ## Análise descritiva dos dados <div style="width: 800px; height: 550px; white-space: nowrap; overflow-x: scroll; overflow-y: scroll; border: 0; padding: 0px; display: inline-block;"> ```r par(mfrow=c(2,3)) boxplot(dados$Temperatura ~ dados$ponto, ylab="Temperatura", xlab = "ponto", cex.lab = 1.5) boxplot(dados$pH ~ dados$ponto, ylab="pH", xlab = "ponto", cex.lab = 1.5) boxplot(dados$Potêncial.de.oxirredução ~ dados$ponto, ylab = "Potêncial de oxirredução", xlab = "ponto", cex.lab = 1.5) boxplot(dados$Turbidez ~ dados$ponto, ylab="Turbidez", xlab = "ponto", cex.lab = 1.5) boxplot(dados$Oxigênio.dissolvido ~ dados$ponto, ylab = "Oxigênio dissolvido", xlab = "ponto", cex.lab = 1.5) boxplot(dados$Espécie.B ~ dados$ponto, ylab = "Espécie B", xlab = "ponto", cex.lab = 1.5) ``` 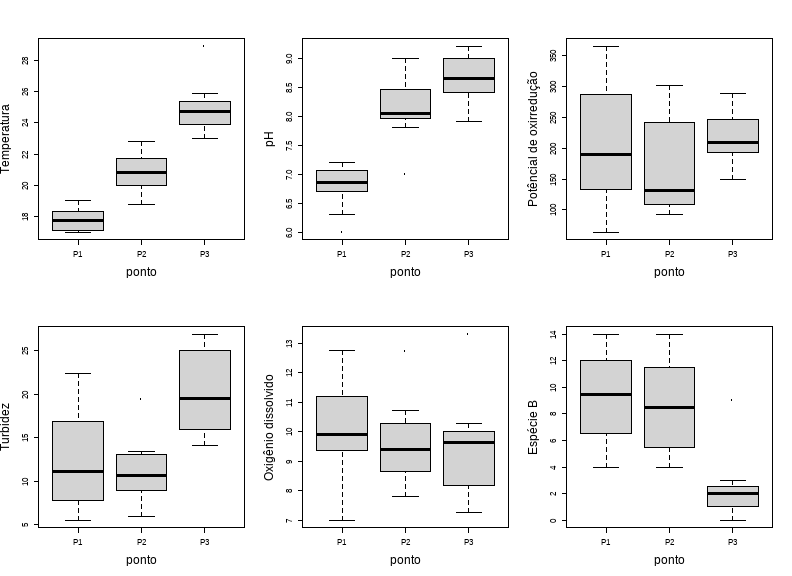<!-- --> --- # PCA no R ## A Análise de Componentes Principais Rodar a análise e salvar no objeto "pca.dados" ```r pca.dados = princomp(dados[,-1], cor=T) ``` ## Resultados da análise PCA ### Visualiza a proporção da variância total explicativa de cada componente principal. - Standard deviation = Autovalor - Proportion of Variance = o quanto cada componente explica a variação dos dados - Cumulative Proportion = % acumulada de explicabilidade de todos os fatores <div style="width: 800px; height: 280px; white-space: nowrap; overflow-x: scroll; overflow-y: scroll; border: 0; padding: 0px; display: inline-block;"> ```r summary(pca.dados) ``` ``` ## Importance of components: ## Comp.1 Comp.2 Comp.3 Comp.4 Comp.5 ## Standard deviation 1.6867171 1.1400420 0.9888787 0.9138858 0.75354753 ## Proportion of Variance 0.4064307 0.1856708 0.1396973 0.1193125 0.08111913 ## Cumulative Proportion 0.4064307 0.5921015 0.7317988 0.8511112 0.93223037 ## Comp.6 Comp.7 ## Standard deviation 0.51295608 0.45963403 ## Proportion of Variance 0.03758913 0.03018049 ## Cumulative Proportion 0.96981951 1.00000000 ``` --- # PCA no R ## Resultados da análise PCA ### Cargas fatoriais Coeficientes das combinações lineares das variáveis contínuas. ```r pca.dados$loadings ``` ``` ## ## Loadings: ## Comp.1 Comp.2 Comp.3 Comp.4 Comp.5 Comp.6 Comp.7 ## Temperatura 0.513 0.136 0.283 0.246 0.228 0.724 ## pH 0.469 0.263 0.345 0.405 -0.653 ## Potêncial.de.oxirredução 0.112 -0.699 -0.200 -0.336 0.589 ## Turbidez 0.447 -0.344 -0.558 0.565 -0.207 ## Oxigênio.dissolvido -0.197 -0.460 -0.249 0.802 -0.136 -0.162 ## Espécie.A -0.301 0.933 0.106 -0.148 ## Espécie.B -0.509 0.158 0.166 0.315 0.762 ## ## Comp.1 Comp.2 Comp.3 Comp.4 Comp.5 Comp.6 Comp.7 ## SS loadings 1.000 1.000 1.000 1.000 1.000 1.000 1.000 ## Proportion Var 0.143 0.143 0.143 0.143 0.143 0.143 0.143 ## Cumulative Var 0.143 0.286 0.429 0.571 0.714 0.857 1.000 ``` --- # PCA no R ## Resultados da análise PCA ### Escores padronizados <div style="width: 800px; height: 500px; white-space: nowrap; overflow-x: scroll; overflow-y: scroll; border: 0; padding: 0px; display: inline-block;"> ```r pca.dados$scores ``` ``` ## Comp.1 Comp.2 Comp.3 Comp.4 Comp.5 Comp.6 ## [1,] -2.0143434 -0.76721378 0.63141269 -0.53424415 -0.27883912 -0.14395527 ## [2,] -1.8763275 0.65260152 0.69181931 -0.58252207 -0.18312365 -0.56380364 ## [3,] -1.5630327 -2.14886204 -0.56610836 -0.05427515 0.01665163 -0.36270237 ## [4,] -0.8543748 -2.79315010 -0.87606417 -1.01870927 -0.73374067 0.48303801 ## [5,] -0.2972373 -0.98452386 -0.70192145 -0.86629279 -0.78331502 0.31921099 ## [6,] -1.0922025 -1.34920317 -1.23255848 -1.37508377 1.08086654 -1.04992352 ## [7,] -0.6055455 1.44046822 -0.43089677 -2.02586934 -1.88341436 -0.04182790 ## [8,] -2.3762894 0.77012339 1.20494986 1.03892756 -0.58659460 -0.57026348 ## [9,] -1.3622561 -0.06226923 1.47345803 0.09972727 -0.90831802 0.74446830 ## [10,] -2.1223415 -1.76404160 1.15935131 0.34364435 0.35376498 0.14629683 ## [11,] -2.5268365 -1.33739002 1.49889558 1.60258865 -0.36376063 0.12216651 ## [12,] -2.2909307 0.61672288 1.77477157 -1.20370144 0.15760064 -0.13753962 ## [13,] -0.6687743 0.94458884 -1.53157774 0.61052169 -0.73025126 -0.24836172 ## [14,] -0.5326564 0.71037292 -0.57272600 -0.43940499 -0.74491232 -0.33641706 ## [15,] 0.9411748 -0.17877390 -1.34863425 0.02306394 0.02901103 -0.02434132 ## [16,] -1.0830905 0.31937339 -0.27218348 -0.68531786 1.13232273 0.24759576 ## [17,] -1.0220832 -0.10223227 -0.46015338 -0.21765401 2.06696690 0.22116127 ## [18,] -0.6592473 -0.89869336 -0.38056177 0.32699713 1.34866324 0.98320988 ## [19,] -1.4294603 2.35723653 -1.08645438 0.29696287 0.72059348 0.97143134 ## [20,] -1.1419934 0.87304571 -1.63218781 2.23393446 -0.29000442 0.15944340 ## [21,] -0.8782548 1.55749563 -0.25476552 -0.30420440 0.14618957 0.04243749 ## [22,] -0.1019164 1.26033366 -0.26474307 0.74104171 0.33964846 -0.07880345 ## [23,] 0.1793464 1.77528678 0.33343929 -0.21113344 -0.27755516 0.12316237 ## [24,] 0.5225279 1.58984869 0.78211987 -0.32451701 0.43775349 -0.53921294 ## [25,] 0.8304486 0.34688874 0.68579992 1.28491259 0.17099734 0.75585186 ## [26,] 2.2128771 -0.87068395 -0.01122491 0.03547090 -0.53078076 0.62467005 ## [27,] 1.4684776 -1.35711881 -0.80190592 2.15598628 -0.95643982 -0.27800486 ## [28,] 2.6153130 0.30233987 -0.16675795 -0.88136951 -0.91243092 0.60820078 ## [29,] 1.4780096 -0.27259294 -0.77490076 -0.53088525 0.81998544 -0.45065333 ## [30,] 2.0457488 -0.44041093 -1.14620719 -0.10616303 -0.09343819 0.01567880 ## [31,] 1.2374969 0.43755307 -1.16457610 0.09591129 0.09633336 -0.34512862 ## [32,] 2.3225773 0.22647639 0.85625598 -0.03607216 0.67635081 -0.01600201 ## [33,] 2.1816152 -0.14201965 0.81477995 0.77729356 -0.08819366 -0.88427988 ## [34,] 3.0276162 -0.91815355 1.13543702 -0.77234057 -0.03455581 0.23097201 ## [35,] 3.4592084 0.25254024 1.80879543 -0.32421929 0.48358452 0.43026965 ## [36,] 1.9767566 -0.04596329 0.82582365 0.82699527 0.30238422 -1.15804431 ## Comp.7 ## [1,] 0.490548109 ## [2,] -0.203628989 ## [3,] 0.029149461 ## [4,] 0.515482107 ## [5,] -0.037095065 ## [6,] -0.194827553 ## [7,] -0.234960158 ## [8,] 0.063028384 ## [9,] -0.044467418 ## [10,] -0.181719761 ## [11,] 0.085410426 ## [12,] 0.051815679 ## [13,] -0.076629186 ## [14,] 0.726853302 ## [15,] -1.290961096 ## [16,] -0.528741517 ## [17,] 0.003596358 ## [18,] -0.047393004 ## [19,] 0.319921641 ## [20,] 0.166906639 ## [21,] -0.119840138 ## [22,] -0.737324982 ## [23,] -0.535680296 ## [24,] 0.187432983 ## [25,] 0.242201035 ## [26,] -0.163137302 ## [27,] -0.362869348 ## [28,] 0.053516812 ## [29,] 0.283047926 ## [30,] 0.458840263 ## [31,] 1.150166844 ## [32,] 0.190545596 ## [33,] -0.271677429 ## [34,] -0.947151644 ## [35,] 0.724162232 ## [36,] 0.235479086 ``` --- # PCA no R ## Resultados da análise PCA ### Quantos componentes? **Critério de Broken-stick**: Autovalor > 1. ```r plot(pca.dados, main = "", cex.lab = 1.5) ``` 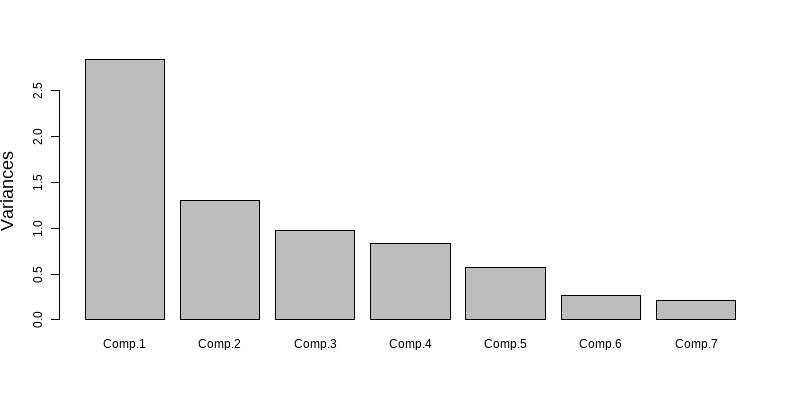<!-- --> --- # PCA no R ## Resultados da análise PCA ### Diagrama de ordenação Representação gráfica da PCA <div style="width: 800px; height: 500px; white-space: nowrap; overflow-x: scroll; overflow-y: scroll; border: 0; padding: 0px; display: inline-block;"> ```r # o comando colour coloriu os pontos amostrados autoplot(pca.dados, data = dados, colour = 'ponto', loadings = TRUE, loadings.colour = 'blue', loadings.label = TRUE, loadings.label.size = 6) + theme(text = element_text(size = 14)) + labs(color = "Ponto de coleta") + xlab("CP 1 (40.64%)") + ylab("CP 2 (18.57%)") ``` 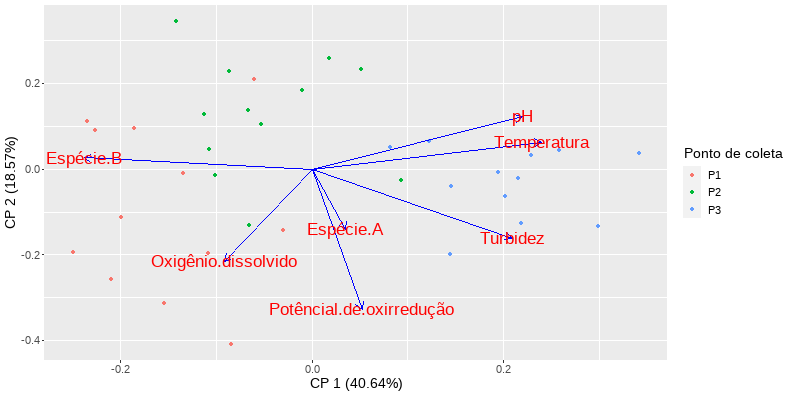<!-- --> --- class: clear background-image: url(fig/Slide13.PNG) background-size: 850px --- .center[ # OBRIGADA!! <img src="https://www.mmfava.com/marilia.png" style="width:30%;"> ## Marília Melo Favalesso <svg viewBox="0 0 512 512" style="height:1em;position:relative;display:inline-block;top:.1em;fill:#000000;" xmlns="http://www.w3.org/2000/svg"> <path d="M502.3 190.8c3.9-3.1 9.7-.2 9.7 4.7V400c0 26.5-21.5 48-48 48H48c-26.5 0-48-21.5-48-48V195.6c0-5 5.7-7.8 9.7-4.7 22.4 17.4 52.1 39.5 154.1 113.6 21.1 15.4 56.7 47.8 92.2 47.6 35.7.3 72-32.8 92.3-47.6 102-74.1 131.6-96.3 154-113.7zM256 320c23.2.4 56.6-29.2 73.4-41.4 132.7-96.3 142.8-104.7 173.4-128.7 5.8-4.5 9.2-11.5 9.2-18.9v-19c0-26.5-21.5-48-48-48H48C21.5 64 0 85.5 0 112v19c0 7.4 3.4 14.3 9.2 18.9 30.6 23.9 40.7 32.4 173.4 128.7 16.8 12.2 50.2 41.8 73.4 41.4z"></path></svg> [mariliabioufpr@gmail.com](mariliabioufpr@gmail.com) <svg viewBox="0 0 496 512" style="height:1em;position:relative;display:inline-block;top:.1em;fill:#000000;" xmlns="http://www.w3.org/2000/svg"> <path d="M248 8C111.03 8 0 119.03 0 256s111.03 248 248 248 248-111.03 248-248S384.97 8 248 8zm82.29 357.6c-3.9 3.88-7.99 7.95-11.31 11.28-2.99 3-5.1 6.7-6.17 10.71-1.51 5.66-2.73 11.38-4.77 16.87l-17.39 46.85c-13.76 3-28 4.69-42.65 4.69v-27.38c1.69-12.62-7.64-36.26-22.63-51.25-6-6-9.37-14.14-9.37-22.63v-32.01c0-11.64-6.27-22.34-16.46-27.97-14.37-7.95-34.81-19.06-48.81-26.11-11.48-5.78-22.1-13.14-31.65-21.75l-.8-.72a114.792 114.792 0 0 1-18.06-20.74c-9.38-13.77-24.66-36.42-34.59-51.14 20.47-45.5 57.36-82.04 103.2-101.89l24.01 12.01C203.48 89.74 216 82.01 216 70.11v-11.3c7.99-1.29 16.12-2.11 24.39-2.42l28.3 28.3c6.25 6.25 6.25 16.38 0 22.63L264 112l-10.34 10.34c-3.12 3.12-3.12 8.19 0 11.31l4.69 4.69c3.12 3.12 3.12 8.19 0 11.31l-8 8a8.008 8.008 0 0 1-5.66 2.34h-8.99c-2.08 0-4.08.81-5.58 2.27l-9.92 9.65a8.008 8.008 0 0 0-1.58 9.31l15.59 31.19c2.66 5.32-1.21 11.58-7.15 11.58h-5.64c-1.93 0-3.79-.7-5.24-1.96l-9.28-8.06a16.017 16.017 0 0 0-15.55-3.1l-31.17 10.39a11.95 11.95 0 0 0-8.17 11.34c0 4.53 2.56 8.66 6.61 10.69l11.08 5.54c9.41 4.71 19.79 7.16 30.31 7.16s22.59 27.29 32 32h66.75c8.49 0 16.62 3.37 22.63 9.37l13.69 13.69a30.503 30.503 0 0 1 8.93 21.57 46.536 46.536 0 0 1-13.72 32.98zM417 274.25c-5.79-1.45-10.84-5-14.15-9.97l-17.98-26.97a23.97 23.97 0 0 1 0-26.62l19.59-29.38c2.32-3.47 5.5-6.29 9.24-8.15l12.98-6.49C440.2 193.59 448 223.87 448 256c0 8.67-.74 17.16-1.82 25.54L417 274.25z"></path></svg> [www.mmfava.com](www.mmfava.com) <svg viewBox="0 0 512 512" style="height:1em;position:relative;display:inline-block;top:.1em;fill:#000000;" xmlns="http://www.w3.org/2000/svg"> <path d="M459.37 151.716c.325 4.548.325 9.097.325 13.645 0 138.72-105.583 298.558-298.558 298.558-59.452 0-114.68-17.219-161.137-47.106 8.447.974 16.568 1.299 25.34 1.299 49.055 0 94.213-16.568 130.274-44.832-46.132-.975-84.792-31.188-98.112-72.772 6.498.974 12.995 1.624 19.818 1.624 9.421 0 18.843-1.3 27.614-3.573-48.081-9.747-84.143-51.98-84.143-102.985v-1.299c13.969 7.797 30.214 12.67 47.431 13.319-28.264-18.843-46.781-51.005-46.781-87.391 0-19.492 5.197-37.36 14.294-52.954 51.655 63.675 129.3 105.258 216.365 109.807-1.624-7.797-2.599-15.918-2.599-24.04 0-57.828 46.782-104.934 104.934-104.934 30.213 0 57.502 12.67 76.67 33.137 23.715-4.548 46.456-13.32 66.599-25.34-7.798 24.366-24.366 44.833-46.132 57.827 21.117-2.273 41.584-8.122 60.426-16.243-14.292 20.791-32.161 39.308-52.628 54.253z"></path></svg> [Twitter: @mmfbee](https://twitter.com/mmfbee) <svg viewBox="0 0 496 512" style="height:1em;position:relative;display:inline-block;top:.1em;fill:#000000;" xmlns="http://www.w3.org/2000/svg"> <path d="M165.9 397.4c0 2-2.3 3.6-5.2 3.6-3.3.3-5.6-1.3-5.6-3.6 0-2 2.3-3.6 5.2-3.6 3-.3 5.6 1.3 5.6 3.6zm-31.1-4.5c-.7 2 1.3 4.3 4.3 4.9 2.6 1 5.6 0 6.2-2s-1.3-4.3-4.3-5.2c-2.6-.7-5.5.3-6.2 2.3zm44.2-1.7c-2.9.7-4.9 2.6-4.6 4.9.3 2 2.9 3.3 5.9 2.6 2.9-.7 4.9-2.6 4.6-4.6-.3-1.9-3-3.2-5.9-2.9zM244.8 8C106.1 8 0 113.3 0 252c0 110.9 69.8 205.8 169.5 239.2 12.8 2.3 17.3-5.6 17.3-12.1 0-6.2-.3-40.4-.3-61.4 0 0-70 15-84.7-29.8 0 0-11.4-29.1-27.8-36.6 0 0-22.9-15.7 1.6-15.4 0 0 24.9 2 38.6 25.8 21.9 38.6 58.6 27.5 72.9 20.9 2.3-16 8.8-27.1 16-33.7-55.9-6.2-112.3-14.3-112.3-110.5 0-27.5 7.6-41.3 23.6-58.9-2.6-6.5-11.1-33.3 2.6-67.9 20.9-6.5 69 27 69 27 20-5.6 41.5-8.5 62.8-8.5s42.8 2.9 62.8 8.5c0 0 48.1-33.6 69-27 13.7 34.7 5.2 61.4 2.6 67.9 16 17.7 25.8 31.5 25.8 58.9 0 96.5-58.9 104.2-114.8 110.5 9.2 7.9 17 22.9 17 46.4 0 33.7-.3 75.4-.3 83.6 0 6.5 4.6 14.4 17.3 12.1C428.2 457.8 496 362.9 496 252 496 113.3 383.5 8 244.8 8zM97.2 352.9c-1.3 1-1 3.3.7 5.2 1.6 1.6 3.9 2.3 5.2 1 1.3-1 1-3.3-.7-5.2-1.6-1.6-3.9-2.3-5.2-1zm-10.8-8.1c-.7 1.3.3 2.9 2.3 3.9 1.6 1 3.6.7 4.3-.7.7-1.3-.3-2.9-2.3-3.9-2-.6-3.6-.3-4.3.7zm32.4 35.6c-1.6 1.3-1 4.3 1.3 6.2 2.3 2.3 5.2 2.6 6.5 1 1.3-1.3.7-4.3-1.3-6.2-2.2-2.3-5.2-2.6-6.5-1zm-11.4-14.7c-1.6 1-1.6 3.6 0 5.9 1.6 2.3 4.3 3.3 5.6 2.3 1.6-1.3 1.6-3.9 0-6.2-1.4-2.3-4-3.3-5.6-2z"></path></svg> [Github: mmfava](https://github.com/mmfava) ]